Program

The Swedish Bioinformatics Workshop (SBW) is an annual event that has been organized by different universities in Sweden since 2000. It is the biggest meeting of bioinformaticians and computational biologists in Sweden. This year, we are happy to announce that the event will take place in Stockholm on November 6-7 at Nya Karolinska Sjukhuset, Solna.
The event is targeted at, and organized by, PhD students and postdocs, but it’s open to everyone working with any kind of bioinformatics or computational biology. Senior scientists and non-academic participants are welcome. In addition, SBW 2023 is part of the Nordic Computational Biology week and is open to participants from other Nordic countries. Register for virtual participation at forms.gle/qvMYdZQTH5oMduhX6 and visit https://bit.ly/ncb-week-2023 for updates on regional programmes.
On this website, we will be updating more information as the arrangements of the workshop proceed.
Registration
While on-site registration is no longer available, you can still register for online participation!
Registration Fees
| Before August 26th 2023 | After August 26th 2023 | |
| Student/Postdoc | Free | Free |
| Other academic participants | 500 SEK | 1000 SEK |
| Non-academic participants | 1500 SEK | 2000 SEK |
Keynote Speakers

A. Murat Eren
University of Oldenburg
Meren’s research explores microbial life through ‘omics strategies and lab experiments, developing computational approaches and open-source software. They aim to unravel how microbes interact, evolve, disperse, and adapt to environmental changes from complex datasets.

Ida-Maria Sintorn
Uppsala University
Ida-Maria Sintorn’s main research interests are image processing for 1) automated content driven multi-scale electron microscopy , and 2) improved explainability and incorporating user guidance in machine learning and pattern recognition tools. Besides theoretical development, her research is focused on biomedical applications such as diagnostics, drug development and disease understanding based on microscopy.

Geir Kjetil Ferkingstad Sandve
University of Oslo
With expertise in gene regulation, omics, epidemiology, and immunology, Geir’s research focuses on understanding how B- and T-cells recognize pathogens. They leverage this knowledge for diagnostics and therapeutics while inspiring methodological advancements in machine learning.

Lars Juhl Jensen
Technical University of Denmark
Lars Juhl Jensen conducts cutting-edge research in protein function prediction, omics integration, and network analysis. As a professor at the University of Copenhagen and scientific advisor of ZS | Intomics, his expertise and contributions in these areas have greatly advanced this field of bioinformatics.

Ida Mommenejad
Microsoft Research NYC
With a background in computational neuroscience, reinforcement learning, and human fMRI, Ida builds and evaluates AI inspired by human brains and behavior, focusing on prefrontal and hippocampal cognitive functions like memory, executive function, navigation, and planning. In addition to evaluating AI agents in xbox games, her recent work evaluates planning ability in large language models (LLMs) and augments LLMs inspired by hippocampus and prefrontal cortex.

Max Karlsson
Pixelgen Technologies
Max obtained his Ph.D. in bioinformatics at KTH Royal Institute of Technology in Stockholm and currently serves as a bioinformatics scientist at Pixelgen Technologies, where he specializes in the analysis and development of methods for Molecular Pixelation data. Molecular Pixelation is an innovative single cell spatial proteomics assay that enables simultaneous measurement of protein abundance, spatial distribution, and colocalization of targeted surface proteins on individual cells. This technique provides a powerful tool for exploring the complex spatial organization of proteins within single cells, opening new avenues for advancements in immune system research
Invited national speakers
Tanja Slotte (Stockholm University)
Marija Cvijovic (University of Gothenburg)
Laura Carroll (Umeå University)
Marta Carroni (Stockholm University)
Program
| Time | Monday, November 6th 2023 |
| 10:00-10:15 | Opening remarks |
| 10:15-11:30 | Session 1: Deciphering how adaptive immune cells recognise pathogens: gathering suited data, defining appropriate assessments and incrementally improving machine learning methodology Geir Kjetil Ferkingstad Sandve Machine learning approaches for novel secondary metabolite discovery Laura Carroll Nanometa Live: A User-friendly Interface for Real-time Metagenomic Data Analysis and Pathogen Identification Kristoffer Sandås Alignment-free identification of antibiotic resistance genes Juan Inda |
| 11:30-12:00 | Break |
| 12:00-13:15 | Session 2: Adapting to reality- tools to incorporate a human-in-the-loop in biomedical image based deep learning Ida-Maria Sintorn Enzyme engineering to accelerate the Calvin-Benson-Bessham cycle in cyanobacteria Ute Hoffmann WebSTR: a population-wide database of short tandem repeat variation in humans Oxana Lundström Factors influencing the horizontal transfer of antibiotic resistance genes David Lund |
| 13:15-14:45 | Lunch Break / Career Lunch |
| 14:45-16:00 | Session 3: Lessons from yeast: synergistic effects of damage accumulation, nutrient signalling and metabolism in the context of cellular rejuvenation and healthspan Marija Cvijovic Molecular Pixelation: Spatial proteomics of single-cells by next generation sequencing Max Karlsson Optimal transport model of cell differentiation infers developmental trajectories in murine hematopoiesis data Magnus Tronstad ADMExtract: a tool for rapid mining and comparative analysis of proteomic datasets Rasmus Hammar |
| 16:00-16:30 | Break |
| 16:30-18:30 | Poster Session |
| 18:30-22:00 | Social / Dinner |
| Time | Tuesday, November 7th 2023 |
| 09:00-11:30 | Parallel Workshop |
| 11:30-12:00 | Coffee Break |
| 12:00-13:00 | Session4: Resistant and vulnerable motor neurons show unique temporal gene regulation in SOD1G93A ALS Irene Mei A generalized benchmark for all types of enrichment analysis methods Davide Buzzao Bioinformatics of cryo-EM data analysis: getting the most out of molecules’ images and combining them with prediction methods Marta Carroni Specifying cellular context of regulons for exploring transcriptome-derived gene regulatory networks Mariia Minaeva |
| 13:00-14:15 | Lunch |
| 14:15-15:15 | Session 5: Writing code for those who are looking for a kitchen in a world of restaurants A. Murat Eren Genomic analyses of the Linum distyly supergene reveal convergent evolution at the molecular level Tanja Slotte Neonatal gut Bifidobacterium associates with indole-3-lactic acid levels in blood and risk of ADHD development Michael Widdowson |
| 15:15-15:45 | Coffee Break |
| 15:45-16:45 | Session 6: Decoding Disease Mechanisms: Representational Learning from Multi-Tissue Healthy Human RNA-seq Data such that Latent Space Arithmetics Extracts Disease Modules Hendrik de Weerd Neuroscience-inspired evaluation and architecture for generative AI Ida Momennejad Cost-Reduced Genotyping of SNPs in Large Populations from Pooled Experiments Camille Clouard |
| 16:45-17:00 | Closing remarks |
Workshop Topics
Data Management
The data management workshop will provide participants with the knowledge and skills to effectively manage scientific data. The workshop covers various aspects of data management:
1. general principles and best practices
2. data cleaning and refinement
3. different data management plans
It also provides an opportunity for discussions about data life cycles and what the SciLifeLab Data Centre can offer in terms of data management services.
Multi-omics
The multi-omics workshop focuses on the integration and analysis of data from different types of omics, like genomics, transcriptomics, proteomics, and metabolomics. The workshop aims to provide participants with an understanding of different approaches for integrating multi-omics data, as well as the tools and techniques used in the analysis of these complex datasets. The principle learning objectives of the workshop are:
1. common issues of integrating multidimensional omics data
2. methods of data integration through supervised and unsupervised machine learning
3. understanding how biological network analysis can assist in identifying coordinated patterns between features and how feature communities are associated with phenomic and biological functions
Depending on time, the workshop will conclude with a short hands-on session
Imaging analysis
During this workshop in image analysis, you will learn to:
1. explain fundamental notions of image analysis, such as digitization, image enhancement, segmentation, feature extraction and classification;
2. use basic open source software to apply and evaluate algorithms for solving image analysis problems;
3. understand the basics for planing the steps necessary to solve a realistic image analysis problem;
4. give examples of applications in microscopy-based research where image analysis, including AI-based methods, may or may not provide a feasible solution.
Software development
This workshop on Productive Programming will cover a few key professional software development practices, adapted to a research setting. The aim is to enable you to be more productive and achieve a higher standard of quality and reproducibility in your work, while at the same time reducing development effort.
Main topics covered:
1. Modular design
2. Testing on a tight schedule: correctness and reproducibility
3. Agile development
The content is aimed both at beginner programmers, as well as more advanced coders with an informal computational background. The methods can be easily used in most programming languages, though examples will be in Python and/or R.
Venue
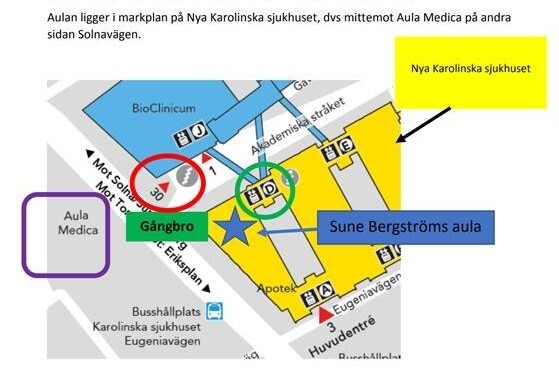
The lectures will be held at Sune Bergström’s Aula at Karolinska Sjukhuset (Solnavägen 30, Stockholm)
The lecture venue can be easily reached with the public transportation with buses 3,6, and 77. Tickets to the bus can be bought via SL app or paying directly with the debit card to the card reader monitor inside the buses. For more information and planning the travels within Stockholm, check sl.se. For more information about parking around Karolinska Sjukhuset, check the website of Karolinska Sjukhuset .
Organization Committee

From left to right: María Bueno Álvez, Philipp Rentzsch, Katja Kozjek, Marcel Tarbier, Kristine Bilgrav Saether, Nilay Peker, Lauri Mesilaakso, Alejandro Rodríguez Gijón and Hauke Wernecke. Missing from photo: Vaishnovi Sekar and Emilia Lahtinen

Kristine Bilgrav Saether
PhD Student at Karolinska Institutet (Rare Diseases group, Stockholm).
Genomics of rare diseases
Maria Bueno Álvez
PhD Student at KTH Royal Institute of Technology & Science for Life Laboratory (Stockholm)
Disease signatures in plasma proteomics data from patients of cancer, cardiovascular conditions, among others.
Twitter: @_buenoalvez


Katja Kozjek
Postdoctoral researcher at Karolinska Institutet (Centre for Translational Microbiome
Research, Stockholm).
Association of gut microbiome with different cancer types.
Linkedin: @katja-kozjek
Emilia Lahtinen
PhD Student at Gedea Biotech AB (Lund) and Centre for Translational Microbiome Research, Karolinska Institutet (Stockholm).
Impact of novel antibiotics for bacterial vaginosis on the vaginal microbiome.
Linkedin: @emilia-lahtinen


Alejandro Rodríguez Gijón
PhD Student at Stockholm University and SciLifeLab (Sarahi Garcia’s group).
Comparative genomics of Archaea and Bacteria in aquatic microbial ecology.
Twitter: @_RodriguezGijon
Github: @alejandrorgijon
Vaishnovi Sekar
PhD Student at Stockholm University and SciLifeLab.
MicroRNA biology and single cells.
Twitter: @vaishnovi_sekar


Marcel Tarbier
Postdoc at Karolinska Institute and SciLifeLab (Vincent Pelenchano’s group).
Development of computational tools to impute complex cell features from molecular profiling data (single-cell and multi-omics data).
Mastodon: @MTarbier
LinkedIn: @marcel-tarbier
Lauri Mesilaakso
Systems developer and affiliation is Centre for Translational Microbiome Research (CTMR), KI (Stockholm).
Development of Biobank application for the National Pandemic Centre and support researchers at CTMR.

Nilay Peker
Postdoctoral bioinformatician at Karolinska Institutet (Centre for Translational Microbiome Research, Stockholm).
Microbiome study of women with recurrent pregnancy loss.
Linkedin: @nilaypeker
Scientific Advisory Board
Arne Elofsson (Stockholm University),
Olof Emanuelsson (KTH),
Olga Dethlefsen (NBIS),
Fredrik Boulund (Karolinska Institute),
Kristoffer Sahlin (Stockholm University),
Carolina Wahlby (Uppsala University),
Anders Andersson(KTH)









